!pip install pandas matplotlib zarr fsspec s3fs intake intake_xarray intake_parquet
Show code cell output
Requirement already satisfied: pandas in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (2.2.2)
Requirement already satisfied: matplotlib in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (3.9.0)
Requirement already satisfied: zarr in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (2.18.2)
Requirement already satisfied: fsspec in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (2024.6.0)
Requirement already satisfied: s3fs in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (2024.6.0)
Requirement already satisfied: intake in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (2.0.5)
Requirement already satisfied: intake_xarray in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (0.7.0)
Requirement already satisfied: intake_parquet in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (0.3.0)
Requirement already satisfied: numpy>=1.23.2 in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from pandas) (1.26.4)
Requirement already satisfied: python-dateutil>=2.8.2 in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from pandas) (2.9.0)
Requirement already satisfied: pytz>=2020.1 in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from pandas) (2024.1)
Requirement already satisfied: tzdata>=2022.7 in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from pandas) (2024.1)
Requirement already satisfied: contourpy>=1.0.1 in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from matplotlib) (1.2.1)
Requirement already satisfied: cycler>=0.10 in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from matplotlib) (0.12.1)
Requirement already satisfied: fonttools>=4.22.0 in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from matplotlib) (4.53.0)
Requirement already satisfied: kiwisolver>=1.3.1 in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from matplotlib) (1.4.5)
Requirement already satisfied: packaging>=20.0 in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from matplotlib) (24.1)
Requirement already satisfied: pillow>=8 in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from matplotlib) (10.3.0)
Requirement already satisfied: pyparsing>=2.3.1 in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from matplotlib) (3.1.2)
Requirement already satisfied: asciitree in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from zarr) (0.3.3)
Requirement already satisfied: numcodecs>=0.10.0 in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from zarr) (0.12.1)
Requirement already satisfied: fasteners in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from zarr) (0.19)
Requirement already satisfied: aiobotocore<3.0.0,>=2.5.4 in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from s3fs) (2.13.0)
Requirement already satisfied: aiohttp!=4.0.0a0,!=4.0.0a1 in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from s3fs) (3.9.5)
Requirement already satisfied: pyyaml in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from intake) (6.0.1)
Requirement already satisfied: appdirs in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from intake) (1.4.4)
Requirement already satisfied: xarray>=02022 in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from intake_xarray) (2024.5.0)
Requirement already satisfied: dask>=2.2 in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from intake_xarray) (2024.5.2)
Requirement already satisfied: netcdf4 in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from intake_xarray) (1.6.5)
Requirement already satisfied: msgpack in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from intake_xarray) (1.0.8)
Requirement already satisfied: requests in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from intake_xarray) (2.32.3)
Requirement already satisfied: fastparquet in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from intake_parquet) (2024.5.0)
Requirement already satisfied: pyarrow in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from intake_parquet) (16.1.0)
Requirement already satisfied: botocore<1.34.107,>=1.34.70 in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from aiobotocore<3.0.0,>=2.5.4->s3fs) (1.34.106)
Requirement already satisfied: wrapt<2.0.0,>=1.10.10 in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from aiobotocore<3.0.0,>=2.5.4->s3fs) (1.16.0)
Requirement already satisfied: aioitertools<1.0.0,>=0.5.1 in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from aiobotocore<3.0.0,>=2.5.4->s3fs) (0.11.0)
Requirement already satisfied: aiosignal>=1.1.2 in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from aiohttp!=4.0.0a0,!=4.0.0a1->s3fs) (1.3.1)
Requirement already satisfied: attrs>=17.3.0 in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from aiohttp!=4.0.0a0,!=4.0.0a1->s3fs) (23.2.0)
Requirement already satisfied: frozenlist>=1.1.1 in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from aiohttp!=4.0.0a0,!=4.0.0a1->s3fs) (1.4.1)
Requirement already satisfied: multidict<7.0,>=4.5 in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from aiohttp!=4.0.0a0,!=4.0.0a1->s3fs) (6.0.5)
Requirement already satisfied: yarl<2.0,>=1.0 in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from aiohttp!=4.0.0a0,!=4.0.0a1->s3fs) (1.9.4)
Requirement already satisfied: click>=8.1 in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from dask>=2.2->intake_xarray) (8.1.7)
Requirement already satisfied: cloudpickle>=1.5.0 in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from dask>=2.2->intake_xarray) (3.0.0)
Requirement already satisfied: partd>=1.2.0 in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from dask>=2.2->intake_xarray) (1.4.2)
Requirement already satisfied: toolz>=0.10.0 in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from dask>=2.2->intake_xarray) (0.12.1)
Requirement already satisfied: importlib-metadata>=4.13.0 in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from dask>=2.2->intake_xarray) (7.1.0)
Requirement already satisfied: six>=1.5 in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from python-dateutil>=2.8.2->pandas) (1.16.0)
Requirement already satisfied: cramjam>=2.3 in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from fastparquet->intake_parquet) (2.8.3)
Requirement already satisfied: cftime in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from netcdf4->intake_xarray) (1.6.4)
Requirement already satisfied: certifi in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from netcdf4->intake_xarray) (2024.6.2)
Requirement already satisfied: charset-normalizer<4,>=2 in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from requests->intake_xarray) (3.3.2)
Requirement already satisfied: idna<4,>=2.5 in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from requests->intake_xarray) (3.7)
Requirement already satisfied: urllib3<3,>=1.21.1 in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from requests->intake_xarray) (2.2.1)
Requirement already satisfied: jmespath<2.0.0,>=0.7.1 in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from botocore<1.34.107,>=1.34.70->aiobotocore<3.0.0,>=2.5.4->s3fs) (1.0.1)
Requirement already satisfied: zipp>=0.5 in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from importlib-metadata>=4.13.0->dask>=2.2->intake_xarray) (3.19.2)
Requirement already satisfied: locket in /Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages (from partd>=1.2.0->dask>=2.2->intake_xarray) (1.0.0)
Tutorial: Accessing Data with Intake & S3#
This notebooks demonstrates how to remotely load data from the archive, stored in a s3 bucket, using intake.
import intake
import matplotlib.pyplot as plt
import pandas as pd
import yaml
import PIL
from IPython.display import Image
Accessing the Shot Index#
Before we can load data from the archive, we find the url where the data is located.
To do this we will use a intake catalog. intake catalogs are a way of abstracting how data is loaded from the user. intake is means that you don’t need to know the details of where the data is stored or how to read it.
To open the catalog we can use intake.open_catalog and give it the path to where our catalog is hosted.
The outpt shows that the catalog contains two sources: index and shots
indexis a source that reads metadata about different objects in the archive. It provides an index of different data objects stored in the archive.level1is a source that reads from about level1 sources, which contain data directly from the tokamak. In the future, derived sources will be added at other product levels.
catalog = intake.open_catalog('https://mastapp.site/intake/catalog.yml')
list(catalog)
['index', 'level1']
Let’s look at the index. The index also contains different product levels. For now, we are only interested in level1 products.
list(catalog.index)
['level1']
Lets use the index source to read in metadata about all the different shots.
Below we read the shot metadata (stored as JSON) directly into a pandas dataframe. The output is a table of metadata including urls for each shot in the archive.
df = pd.DataFrame(catalog.index.level1.shots.read())
df = df[['shot_id', 'campaign', 'url']]
df
| shot_id | campaign | url | |
|---|---|---|---|
| 0 | 11695 | M5 | s3://mast/level1/shots/11695.zarr |
| 1 | 11696 | M5 | s3://mast/level1/shots/11696.zarr |
| 2 | 11697 | M5 | s3://mast/level1/shots/11697.zarr |
| 3 | 11698 | M5 | s3://mast/level1/shots/11698.zarr |
| 4 | 11699 | M5 | s3://mast/level1/shots/11699.zarr |
| ... | ... | ... | ... |
| 15548 | 30467 | M9 | s3://mast/level1/shots/30467.zarr |
| 15549 | 30468 | M9 | s3://mast/level1/shots/30468.zarr |
| 15550 | 30469 | M9 | s3://mast/level1/shots/30469.zarr |
| 15551 | 30470 | M9 | s3://mast/level1/shots/30470.zarr |
| 15552 | 30471 | M9 | s3://mast/level1/shots/30471.zarr |
15553 rows × 3 columns
Using the urls of the shots we can load data from the archive.
In the next cell we use the url of the first shot to remotely open data from the amc diagnostic.
intake returns a xr.Dataset object containing all the data for this diagnostic.
shot = df.loc[df.shot_id == 30420].iloc[0]
dataset = catalog.level1.shots(url=shot.url, group='amc').to_dask()
dataset
/Users/rt2549/miniconda3/envs/mast-book/lib/python3.11/site-packages/intake_xarray/base.py:21: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
'dims': dict(self._ds.dims),
<xarray.Dataset> Size: 5MB
Dimensions: (time: 30000)
Coordinates:
* time (time) float32 120kB -2.0 -2.0 -2.0 ... 3.999 4.0 4.0
Data variables: (12/46)
efps_current (time) float32 120kB dask.array<chunksize=(30000,), meta=np.ndarray>
error_field_02 (time) float32 120kB dask.array<chunksize=(30000,), meta=np.ndarray>
error_field_05 (time) float32 120kB dask.array<chunksize=(30000,), meta=np.ndarray>
p2il_coil_current (time) float32 120kB dask.array<chunksize=(30000,), meta=np.ndarray>
p2il_feed_current (time) float32 120kB dask.array<chunksize=(30000,), meta=np.ndarray>
p2iu_coil_current (time) float32 120kB dask.array<chunksize=(30000,), meta=np.ndarray>
... ...
p6u_current (time) float32 120kB dask.array<chunksize=(30000,), meta=np.ndarray>
plasma_current (time) float32 120kB dask.array<chunksize=(30000,), meta=np.ndarray>
sol_current (time) float32 120kB dask.array<chunksize=(30000,), meta=np.ndarray>
status float32 4B ...
tf_current (time) float32 120kB dask.array<chunksize=(30000,), meta=np.ndarray>
version float32 4B ...
Attributes:
description: Plasma Current and PF/TF Coil Currents
file_name: amc0304.20
format: IDA3
mds_name: None
name: amc
quality: Not Checked
shot_id: 30420
signal_type: Analysed
source: amc
uda_name: AMC
uuid: 01aad0c4-2a84-59e2-8b1b-168b4bd66aa3
version: 0Data Analysis with Remote Data#
We’re going to perform a simple plotting task. We will:
Get the URLs for 10 shots in a given range
Load the plasma current data as a
xarray.DatasetSlice every shot between 0 seconds and .3 seconds.
df = df.loc[(df.shot_id <= 30420) & (df.shot_id >= 30410)]
plasma_shots = []
for index, row in df.iterrows():
dataset = catalog.level1.shots(url=row['url'], group='amc')
dataset = dataset.to_dask()
dataset = dataset['plasma_current']
dataset = dataset.sel(time=slice(0, .3))
plasma_shots.append(dataset)
In the code above, we load each item as an xarray dataset, with the data, time, and error data all together.
plasma_shots[0]
<xarray.DataArray 'plasma_current' (time: 1500)> Size: 6kB
dask.array<getitem, shape=(1500,), dtype=float32, chunksize=(1500,), chunktype=numpy.ndarray>
Coordinates:
* time (time) float32 6kB 0.0001998 0.0003998 0.0005996 ... 0.2998 0.3
Attributes: (12/18)
description: Plasma Current
dims: ['time']
file_name: None
format: None
label: Plasma Current
mds_name: \TOP.ANALYSED.AMC.PLASMA:CURRENT
... ...
source: amc
time_index: 0
uda_name: AMC_PLASMA CURRENT
units: kA
uuid: 04b71d20-1e39-538d-9626-a6ef7926a84e
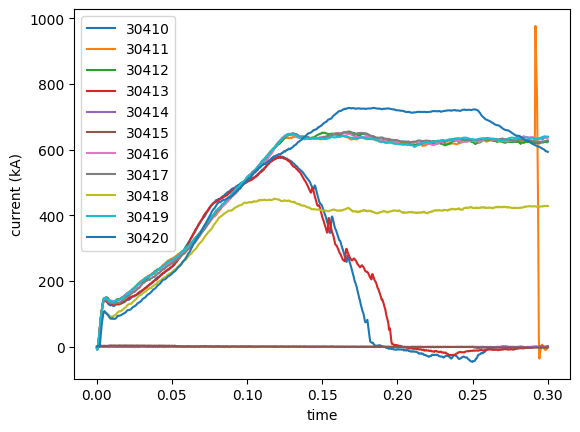
version: 0Finally, we can plot the 10 shots we loaded and cropped.
for current in plasma_shots:
plt.plot(current.time, current.data, label=current.attrs['shot_id'])
plt.xlabel('time')
plt.ylabel(f"current ({current.attrs['units']})")
plt.legend()
<matplotlib.legend.Legend at 0x35bc46110>
Larger Data - Loading RBB Image Data#
In this example we show how to load Image data remotely. Image data are just grouped by source, such as the rbb data. In this example we load all the image data from an rbb group and create a GIF of the contents.
dataset = catalog.level1.shots(url=shot.url, group='rbb')
dataset = dataset.read()
dataset
<xarray.Dataset> Size: 82MB
Dimensions: (time: 286, height: 448, width: 640)
Coordinates:
* time (time) float64 2kB 1.6e-05 0.002016 0.004016 ... 0.308 0.309 0.31
Dimensions without coordinates: height, width
Data variables:
data (time, height, width) uint8 82MB 0 0 2 0 0 0 1 2 ... 2 2 0 4 0 2 0
Attributes: (12/48)
CLASS: IMAGE
IMAGE_SUBCLASS: IMAGE_INDEXED
IMAGE_VERSION: 1.2
board_temp: 0.0
bottom: 680
camera:
... ...
units: pixels
uuid: 10ed506a-3ac4-5e62-8a6b-25a7abfc3171
vbin: 0
version: -1
view: photron HM10 + Dalpha filter
width: 640imgs = [PIL.Image.fromarray(img) for img in dataset.data.values]
imgs[0].save("array.gif", save_all=True, append_images=imgs[1:], duration=50, loop=0)
Image(open('array.gif','rb').read())


