!pip install matplotlib pandas pyarrow seaborn
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
plt.rcParams["font.family"] = "sans"
plt.rcParams["font.size"] = 8
sns.set_palette('muted')
Shot Metadata#
This notebook contains a demonstration of plotting several of the summary statistics that accompany the shot metadata.
Firstly, we’re going to load all the shot data into a pandas dataframe:
summary = pd.read_parquet('https://mastapp.site/parquet/level2/shots')
summary
| shot_id | uuid | equi_max_li3 | ohmnic_max_heating | generic_max_energy_time | generic_min_q95_time | endpoint_url | generic_max_geo_major_radius_time | timestamp | shot_postshot_comment | ... | nbi_power_max_ss | nbi_energy_ss_max_power | nbi_max_ss_power_time | nbi_power_ss_max_current | nbi_power_truby_ss | scenario | rad_o2ratio | radii_c2ratio | shot_scenario | shot_flat_top_duration | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 11766 | 7da2590b-6b7c-5a4e-8125-8b0c38a7c8bf | 1.520526 | 1.038769 | 0.037496 | 0.270 | https://s3.echo.stfc.ac.uk | 0.080 | 2005-01-13 12:02:00 | GOOD PLASMA, RAN FINE. SL JOINT ALARMS RATHER ... | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | None | NaN |
| 1 | 11767 | 594cabe7-8b0a-54a2-9e1e-8b250a858acd | 1.217211 | 1.105280 | 0.019210 | 0.245 | https://s3.echo.stfc.ac.uk | 0.080 | 2005-01-13 12:17:00 | OK BUT LOST VERTICAL CONTROL - FA2 JUST DIED A... | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | None | NaN |
| 2 | 11768 | 23a8941d-e073-5073-8f5e-e53230f55b43 | 1.262946 | 0.849237 | 0.036312 | 0.150 | https://s3.echo.stfc.ac.uk | 0.080 | 2005-01-13 13:30:00 | OK. GOT FA4 BUT NOT FA3 | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | None | NaN |
| 3 | 11769 | e12564d4-f0af-5c8d-b15c-ab59f11144b3 | 1.519628 | 2.149353 | 0.057099 | 0.290 | https://s3.echo.stfc.ac.uk | 0.290 | 2005-01-13 13:44:00 | SLIDING JOINT ALARMS A BIT LOWER. PLASMA OK. | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | None | NaN |
| 4 | 11771 | 1562346b-2097-5d27-8649-07eeb48dc5f7 | 1.788729 | 0.824824 | 0.052359 | 0.285 | https://s3.echo.stfc.ac.uk | 0.260 | 2005-01-13 14:33:00 | GOOD PLASMA F/B CONTROL. SLIDING JOINT ALARMS ... | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | None | NaN |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 11568 | 30467 | db7bc878-869c-59c6-bcc6-cb28cba69cf9 | 6.694992 | NaN | -1.000000 | 0.030 | https://s3.echo.stfc.ac.uk | 0.165 | 2013-09-27 14:03:00 | 'Two times lower DD neutron rate than referenc... | ... | 2.080261 | 70.840835 | 0.12160 | 2.180491 | NaN | 3.0 | NaN | NaN | S6 | NaN |
| 11569 | 30468 | 696b1a86-1986-59a4-8cd8-f968dba4ece4 | 8.221705 | NaN | -1.000000 | 0.030 | https://s3.echo.stfc.ac.uk | 0.155 | 2013-09-27 14:21:00 | 'Good beam.Good repeat.' | ... | 2.083544 | 70.921546 | 0.13065 | 1.985447 | NaN | 2.0 | NaN | NaN | S8 | NaN |
| 11570 | 30469 | dcc5091c-ba43-599e-9c63-f8cb41494a60 | 2.542882 | NaN | -1.000000 | 0.195 | https://s3.echo.stfc.ac.uk | 0.170 | 2013-09-27 14:39:00 | 'Good shot. Modes present.' | ... | 2.163367 | 75.050461 | 0.19560 | 2.288206 | NaN | 3.0 | NaN | NaN | S6 | NaN |
| 11571 | 30470 | 63ccd361-c339-5403-abf3-dec0ad9d4811 | 11.771064 | 10.391692 | 0.008542 | 0.035 | https://s3.echo.stfc.ac.uk | 0.170 | 2013-09-27 15:03:00 | 'No HF gas.' | ... | 2.204641 | 75.045603 | 0.18445 | 2.107413 | NaN | 2.0 | NaN | NaN | S8 | NaN |
| 11572 | 30471 | 0916ffb1-fece-5d3d-81a9-b0504f49929f | 59.918600 | 73.787904 | 0.021805 | 0.035 | https://s3.echo.stfc.ac.uk | 0.125 | 2013-09-27 15:20:00 | 'Good shot.' | ... | 2.062899 | 71.177179 | 0.12170 | NaN | NaN | 2.0 | NaN | NaN | S8 | NaN |
11573 rows × 189 columns
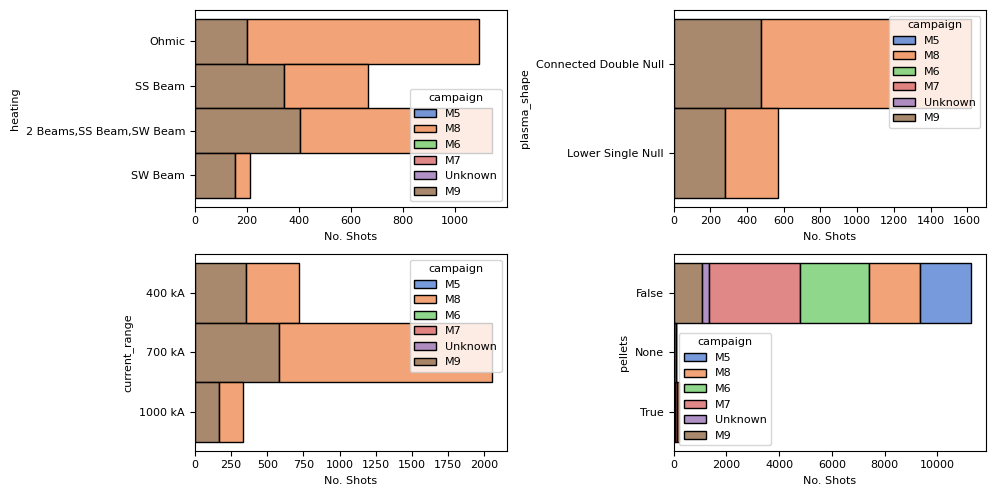
Summary Statistics About Shots#
Let’s look at a summary of simple counts of different shot metadata.
fig, axes = plt.subplots(2, 2, figsize=(10, 5))
ax1, ax2, ax3, ax4 = axes.flatten()
sns.histplot(summary, y='heating', hue='campaign', multiple="stack", ax=ax1)
sns.histplot(summary, y='plasma_shape', hue='campaign', multiple="stack", ax=ax2)
sns.histplot(summary, y='current_range', hue='campaign', multiple="stack", ax=ax3)
sns.histplot(summary, y=summary.pellets.astype(str), hue='campaign', multiple="stack", ax=ax4)
for ax in axes.flatten():
ax.set_xlabel('No. Shots')
plt.tight_layout()
Plasma Beta (\(\beta\)) v.s Confinement Time (\(\tau_E\))#
This plot can show how the efficiency of energy confinement varies with plasma pressure.
plt.figure()
sns.scatterplot(summary, y='cpf_tautot_max', x='cpf_betmhd_max', hue='heating')
plt.xlim(0, 18)
plt.ylim(0, 1)
plt.ylabel('Confinement time $\\tau_E$ (s)')
plt.xlabel('Plasma Beta $\\beta$ (%)')
plt.show()
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
Cell In[5], line 2
1 plt.figure()
----> 2 sns.scatterplot(summary, y='cpf_tautot_max', x='cpf_betmhd_max', hue='heating')
3 plt.xlim(0, 18)
4 plt.ylim(0, 1)
File /srv/fair-mast/.docs-venv/lib/python3.12/site-packages/seaborn/relational.py:615, in scatterplot(data, x, y, hue, size, style, palette, hue_order, hue_norm, sizes, size_order, size_norm, markers, style_order, legend, ax, **kwargs)
606 def scatterplot(
607 data=None, *,
608 x=None, y=None, hue=None, size=None, style=None,
(...) 612 **kwargs
613 ):
--> 615 p = _ScatterPlotter(
616 data=data,
617 variables=dict(x=x, y=y, hue=hue, size=size, style=style),
618 legend=legend
619 )
621 p.map_hue(palette=palette, order=hue_order, norm=hue_norm)
622 p.map_size(sizes=sizes, order=size_order, norm=size_norm)
File /srv/fair-mast/.docs-venv/lib/python3.12/site-packages/seaborn/relational.py:396, in _ScatterPlotter.__init__(self, data, variables, legend)
387 def __init__(self, *, data=None, variables={}, legend=None):
388
389 # TODO this is messy, we want the mapping to be agnostic about
390 # the kind of plot to draw, but for the time being we need to set
391 # this information so the SizeMapping can use it
392 self._default_size_range = (
393 np.r_[.5, 2] * np.square(mpl.rcParams["lines.markersize"])
394 )
--> 396 super().__init__(data=data, variables=variables)
398 self.legend = legend
File /srv/fair-mast/.docs-venv/lib/python3.12/site-packages/seaborn/_base.py:634, in VectorPlotter.__init__(self, data, variables)
629 # var_ordered is relevant only for categorical axis variables, and may
630 # be better handled by an internal axis information object that tracks
631 # such information and is set up by the scale_* methods. The analogous
632 # information for numeric axes would be information about log scales.
633 self._var_ordered = {"x": False, "y": False} # alt., used DefaultDict
--> 634 self.assign_variables(data, variables)
636 # TODO Lots of tests assume that these are called to initialize the
637 # mappings to default values on class initialization. I'd prefer to
638 # move away from that and only have a mapping when explicitly called.
639 for var in ["hue", "size", "style"]:
File /srv/fair-mast/.docs-venv/lib/python3.12/site-packages/seaborn/_base.py:679, in VectorPlotter.assign_variables(self, data, variables)
674 else:
675 # When dealing with long-form input, use the newer PlotData
676 # object (internal but introduced for the objects interface)
677 # to centralize / standardize data consumption logic.
678 self.input_format = "long"
--> 679 plot_data = PlotData(data, variables)
680 frame = plot_data.frame
681 names = plot_data.names
File /srv/fair-mast/.docs-venv/lib/python3.12/site-packages/seaborn/_core/data.py:58, in PlotData.__init__(self, data, variables)
51 def __init__(
52 self,
53 data: DataSource,
54 variables: dict[str, VariableSpec],
55 ):
57 data = handle_data_source(data)
---> 58 frame, names, ids = self._assign_variables(data, variables)
60 self.frame = frame
61 self.names = names
File /srv/fair-mast/.docs-venv/lib/python3.12/site-packages/seaborn/_core/data.py:232, in PlotData._assign_variables(self, data, variables)
230 else:
231 err += "An entry with this name does not appear in `data`."
--> 232 raise ValueError(err)
234 else:
235
236 # Otherwise, assume the value somehow represents data
237
238 # Ignore empty data structures
239 if isinstance(val, Sized) and len(val) == 0:
ValueError: Could not interpret value `cpf_betmhd_max` for `x`. An entry with this name does not appear in `data`.
<Figure size 640x480 with 0 Axes>
Plasma Temperature (\(T_e\)) vs. Plasma Density (\(n_e\))#
This can reveal the relationship between temperature and density, which is critical for achieving the conditions necessary for fusion.
plt.figure()
sns.scatterplot(summary, y='cpf_te0_ipmax', x='cpf_ne0_ipmax', hue='current_range', alpha=0.8)
plt.xlim(0, .8e20)
plt.ylim(0, 1750)
plt.ylabel('Temperature $T_e$ (eV)')
plt.xlabel('Density $n_e$ ($m^{-3}$)')
plt.show()
Plasma Current (\(I_p\)) vs. Confinement Time (\(\tau_E\))#
This can indicate how the plasma current affects the confinement time, providing insights into stability and performance.
plt.figure()
sns.scatterplot(summary, y='cpf_ip_av', x='cpf_tautot_max', hue='current_range', alpha=0.8)
plt.xlim(0, 1)
plt.xlabel('Confinement Time $\\tau_E$ (s)')
plt.ylabel('Average Plasma Current $I_p$ (kA)')
plt.show()

